
原文链接:
https://dx.doi.org/10.1093/nar/gkad295
https://dx.doi.org/10.1093/nar/gkad279
参考文献
1. Liu X, Lu S, Song K, Shen Q, Ni D, Li Q, He X, Zhang H, Wang Q, Chen Y, Li X, Wu J, Sheng C, Chen G, Lu X*, Zhang J*. Unraveling allosteric landscapes of allosterome with ASD. Nucleic Acids Res. 2020, 48(D1): D394-D401.
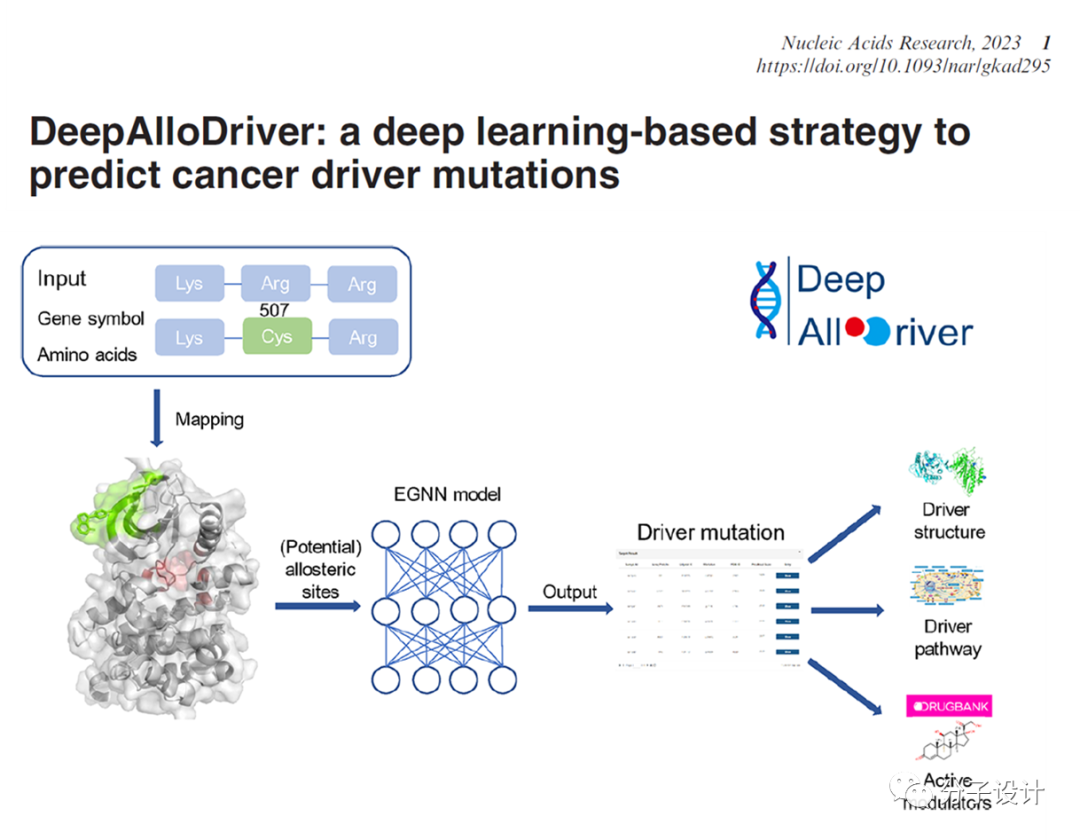
2. Song Q, Li M, Li Q, Lu X, Song K, Zhang Z, Wei J, Zhang L, Wei J, Ye Y, Zha J, Zhang Q, Gao Q, Long J, Liu X, Lu X, Zhang J. DeepAlloDriver: a deep learning-based web strategy to predict cancer driver mutations. Nucleic Acids Res. 2023, doi: 10.1093/nar/gkad295
3. Shen Q, Cheng F, Song H, Lu W, Zhao J, An X, Liu M, Chen G, Zhao Z*, Zhang J*. Proteome-scale investigation of protein allosteric regulation perturbed by somatic mutations in 7,000 cancer genomes. Am J Hum Genet. 2017, 100(1): 5-20.
4. Song K, Li Q, Gao W, Lu S, Shen Q, Liu X, Wu Y, Wang B, Lin H, Chen G, Zhang J*. AlloDriver: a method for the identification and analysis of cancer driver targets. Nucleic Acids Res. 2019, 47(W1): W315-W321.
5. Zha J, Li Q, Liu X, Lin W, Wang T, Wei J, Zhang Z, Lu X, Wu J, Ni D, Song K, Zhang L, Lu X, Lu S, Zhang J. AlloReverse: multiscale understanding among hierarchical allosteric regulations. Nucleic Acids Res. 2023, doi: 10.1093/nar/gkad279
6. Ni D, Wei J, He X, Rehman AU, Li X, Qiu Y, Pu J, Lu S*, Zhang J*. Discovery of cryptic allosteric sites using reversed allosteric communication by a combined computational and experimental strategy. Chem Sci. 2021, 12: 464-476 (Editor’s Choice & ESI Top1% highly cited paper).
本网站所有内容来源注明为“梅斯医学”或“MedSci原创”的文字、图片和音视频资料,版权均属于梅斯医学所有。非经授权,任何媒体、网站或个人不得转载,授权转载时须注明来源为“梅斯医学”。其它来源的文章系转载文章,或“梅斯号”自媒体发布的文章,仅系出于传递更多信息之目的,本站仅负责审核内容合规,其内容不代表本站立场,本站不负责内容的准确性和版权。如果存在侵权、或不希望被转载的媒体或个人可与我们联系,我们将立即进行删除处理。
在此留言